BAT1
| DDX39B | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | DDX39B, BAT1, D6S81E, UAP56, DEAD-box helicase 39B, DExD-box helicase 39B | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 142560; MGI: 99240; HomoloGene: 48376; GeneCards: DDX39B; OMA:DDX39B - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Spliceosome RNA helicase BAT1 is an enzyme that in humans is encoded by the BAT1 gene.[5][6][7]
This gene encodes a member of the DEAD box family of RNA-dependent ATPases that mediate ATP hydrolysis during pre-mRNA splicing. The encoded protein is an essential splicing factor required for association of U2 small nuclear ribonucleoprotein with pre-mRNA, and also plays an important role in mRNA export from the nucleus to the cytoplasm. A cluster of genes, BAT1-BAT5, is localized in the vicinity of the genes for TNF alpha and TNF beta. These genes are all within the human major histocompatibility complex class III region. Mutations in this gene may be associated with rheumatoid arthritis. Alternatively spliced transcript variants encoding the same protein have been described.[7]
References
- ^ a b c ENSG00000225073, ENSG00000237889, ENSG00000229496, ENSG00000215425, ENSG00000235439, ENSG00000225859, ENSG00000230624 GRCh38: Ensembl release 89: ENSG00000198563, ENSG00000225073, ENSG00000237889, ENSG00000229496, ENSG00000215425, ENSG00000235439, ENSG00000225859, ENSG00000230624 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000019432 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Peelman LJ, Chardon P, Nunes M, Renard C, Geffrotin C, Vaiman M, Van Zeveren A, Coppieters W, van de Weghe A, Bouquet Y, et al. (Aug 1995). "The BAT1 gene in the MHC encodes an evolutionarily conserved putative nuclear RNA helicase of the DEAD family". Genomics. 26 (2): 210–8. doi:10.1016/0888-7543(95)80203-X. PMID 7601445.
- ^ Spies T, Bresnahan M, Strominger JL (Dec 1989). "Human major histocompatibility complex contains a minimum of 19 genes between the complement cluster and HLA-B". Proc Natl Acad Sci U S A. 86 (22): 8955–8. Bibcode:1989PNAS...86.8955S. doi:10.1073/pnas.86.22.8955. PMC 298409. PMID 2813433.
- ^ a b "Entrez Gene: BAT1 HLA-B associated transcript 1".
External links
- Human DDX39B genome location and DDX39B gene details page in the UCSC Genome Browser.
- PDBe-KB provides an overview of all the structure information available in the PDB for Human Spliceosome RNA helicase DDX39B (BAT1)
Further reading
- Spies T, Blanck G, Bresnahan M, et al. (1989). "A new cluster of genes within the human major histocompatibility complex". Science. 243 (4888): 214–7. Bibcode:1989Sci...243..214S. doi:10.1126/science.2911734. PMID 2911734.
- Cross SH, Charlton JA, Nan X, Bird AP (1994). "Purification of CpG islands using a methylated DNA binding column". Nat. Genet. 6 (3): 236–44. doi:10.1038/ng0394-236. PMID 8012384. S2CID 12847618.
- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Fleckner J, Zhang M, Valcárcel J, Green MR (1997). "U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction". Genes Dev. 11 (14): 1864–72. doi:10.1101/gad.11.14.1864. PMID 9242493.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, et al. (1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Allcock RJ, Price P, Gaudieri S, et al. (1999). "Characterisation of the human central MHC gene, BAT1: genomic structure and expression". Exp. Clin. Immunogenet. 16 (2): 98–106. doi:10.1159/000019100. PMID 10343160. S2CID 22003689.
- Ota M, Katsuyama Y, Kimura A, et al. (2001). "A second susceptibility gene for developing rheumatoid arthritis in the human MHC is localized within a 70-kb interval telomeric of the TNF genes in the HLA class III region". Genomics. 71 (3): 263–70. doi:10.1006/geno.2000.6371. PMID 11170743.
- Allcock RJ, Williams JH, Price P (2001). "The central MHC gene, BAT1, may encode a protein that down-regulates cytokine production". Genes Cells. 6 (5): 487–94. doi:10.1046/j.1365-2443.2001.00435.x. PMID 11380625. S2CID 10605522.
- Luo ML, Zhou Z, Magni K, et al. (2001). "Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly". Nature. 413 (6856): 644–7. Bibcode:2001Natur.413..644L. doi:10.1038/35098106. PMID 11675789. S2CID 4395388.
- Andersen JS, Lyon CE, Fox AH, et al. (2002). "Directed proteomic analysis of the human nucleolus". Curr. Biol. 12 (1): 1–11. Bibcode:2002CBio...12....1A. doi:10.1016/S0960-9822(01)00650-9. PMID 11790298. S2CID 14132033.
- Strässer K, Masuda S, Mason P, et al. (2002). "TREX is a conserved complex coupling transcription with messenger RNA export". Nature. 417 (6886): 304–8. Bibcode:2002Natur.417..304S. doi:10.1038/nature746. PMID 11979277. S2CID 1112194.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- McCracken S, Longman D, Johnstone IL, et al. (2004). "An evolutionarily conserved role for SRm160 in 3'-end processing that functions independently of exon junction complex formation". J. Biol. Chem. 278 (45): 44153–60. doi:10.1074/jbc.M306856200. PMID 12944400.
- Reuter TY, Medhurst AL, Waisfisz Q, et al. (2003). "Yeast two-hybrid screens imply involvement of Fanconi anemia proteins in transcription regulation, cell signaling, oxidative metabolism, and cellular transport". Exp. Cell Res. 289 (2): 211–21. doi:10.1016/S0014-4827(03)00261-1. PMID 14499622.
- Li J, Hawkins IC, Harvey CD, et al. (2003). "Regulation of alternative splicing by SRrp86 and its interacting proteins". Mol. Cell. Biol. 23 (21): 7437–47. doi:10.1128/MCB.23.21.7437-7447.2003. PMC 207616. PMID 14559993.
- Mungall AJ, Palmer SA, Sims SK, et al. (2003). "The DNA sequence and analysis of human chromosome 6". Nature. 425 (6960): 805–11. Bibcode:2003Natur.425..805M. doi:10.1038/nature02055. PMID 14574404.
- Lehner B, Semple JI, Brown SE, et al. (2004). "Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region". Genomics. 83 (1): 153–67. doi:10.1016/S0888-7543(03)00235-0. PMID 14667819.
- Price P, Wong AM, Williamson D, et al. (2004). "Polymorphisms at positions -22 and -348 in the promoter of the BAT1 gene affect transcription and the binding of nuclear factors". Hum. Mol. Genet. 13 (9): 967–74. doi:10.1093/hmg/ddh113. PMID 15028669.
- v
- t
- e
-
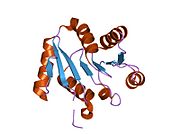 1t5i: Crystal structure of the C-terminal domain of UAP56
1t5i: Crystal structure of the C-terminal domain of UAP56 -
 1t6n: Crystal structure of the N-terminal domain of human UAP56
1t6n: Crystal structure of the N-terminal domain of human UAP56 -
 1xti: Structure of Wildtype human UAP56
1xti: Structure of Wildtype human UAP56 -
 1xtj: structure of human UAP56 in complex with ADP
1xtj: structure of human UAP56 in complex with ADP -
 1xtk: structure of DECD to DEAD mutation of human UAP56
1xtk: structure of DECD to DEAD mutation of human UAP56
 | This enzyme-related article is a stub. You can help Wikipedia by expanding it. |
- v
- t
- e























